Simple BitBIRCH workflow#
This is a tutorial notebook that showcases a simple workflow of BitBIRCH clustering, including a refinement step. If you find this software useful please cite the following articles:
Please reach out to one of the following with any questions or concerns.
Ramon Alain Miranda Quintana:
quintana At chem.ufl.eduKenneth Lopez Perez:
klopezperez At chem.ufl.eduIgnacio Pickering:
ipickering At chem.ufl.eduKrisztina Zsigmond:
kzsigmond At ufl.eduMiroslav Lzicar:
miroslav.lzicar At deepmedchem.com
Set Up#
First lets install the BitBirch-Lean package (if you have not already done so). To do this, run the following commands in your terminal:
git clone https://github.com/mqcomplab/bblean.git
cd bblean
pip install -v .
Lets import the bblean package and some bblean modules, which we will use throughout this example.
[48]:
import bblean
import bblean.plotting as plotting
import bblean.analysis as analysis
Now let’s take some SMILES strings and compute molecular fingerprints:
[49]:
smiles = bblean.load_smiles("./chembl-33-natural-products-subset.smi")
# By default the fps created are of the "ecfp4" kind. Here we use "rdkit"
fps = bblean.fps_from_smiles(smiles, pack=True, n_features=2048, kind="rdkit")
print(f"Shape: {fps.shape}, DType: {fps.dtype}")
Shape: (64086, 256), DType: uint8
The most efficient way to store and manipulate fingerprints is using packed fingerprint arrays. Packed arrays save the features in a compressed representation. To convert between packed and unpacked fingerprints you can use bblean.pack_fingerprints(fps) and bblean.unpack_fingerprints(fps).
[50]:
fps_unpacked = bblean.unpack_fingerprints(fps)
print(f"Shape unpacked: {fps_unpacked.shape}, DType unpacked: {fps_unpacked.dtype}")
fps = bblean.pack_fingerprints(fps_unpacked)
print(f"Shape re-packed: {fps.shape}, DType re-packed: {fps.dtype}")
Shape unpacked: (64086, 2048), DType unpacked: uint8
Shape re-packed: (64086, 256), DType re-packed: uint8
Clustering fingerprints#
Now lets create a BitBirch object, which we can use to cluster the (packed) fingerprints
[51]:
# Initialize the BitBirch tree. In general, diameter is the best merge criterion for
# initial clustering. For more info consult the *refinement* paper.
# 0.5-0.65 is a good threshold range for "rdkit" fingerprints, for ecfp4 use 0.3-0.4 instead
bb_tree = bblean.BitBirch(branching_factor=50, threshold=0.65, merge_criterion="diameter")
# Cluster the packed fingerprints (By default all bblean functions take packed
# fingerprints)
bb_tree.fit(fps)
[51]:
BitBirch(threshold=0.65, branching_factor=50, merge_criterion='diameter')
Finally, lets visualize the clustering results:
[52]:
# First we run a cluster analysis on the resulting ids
clusters = bb_tree.get_cluster_mol_ids()
ca = analysis.cluster_analysis(clusters, fps, smiles)
# Afterwards we can use the utility functions on the bblean.plotting module
plotting.summary_plot(ca, title="Diameter")
# Optionally we can save the cluster analysis metrics as a csv file
ca.dump_metrics("./diameter-metrics.csv")
plt.show()
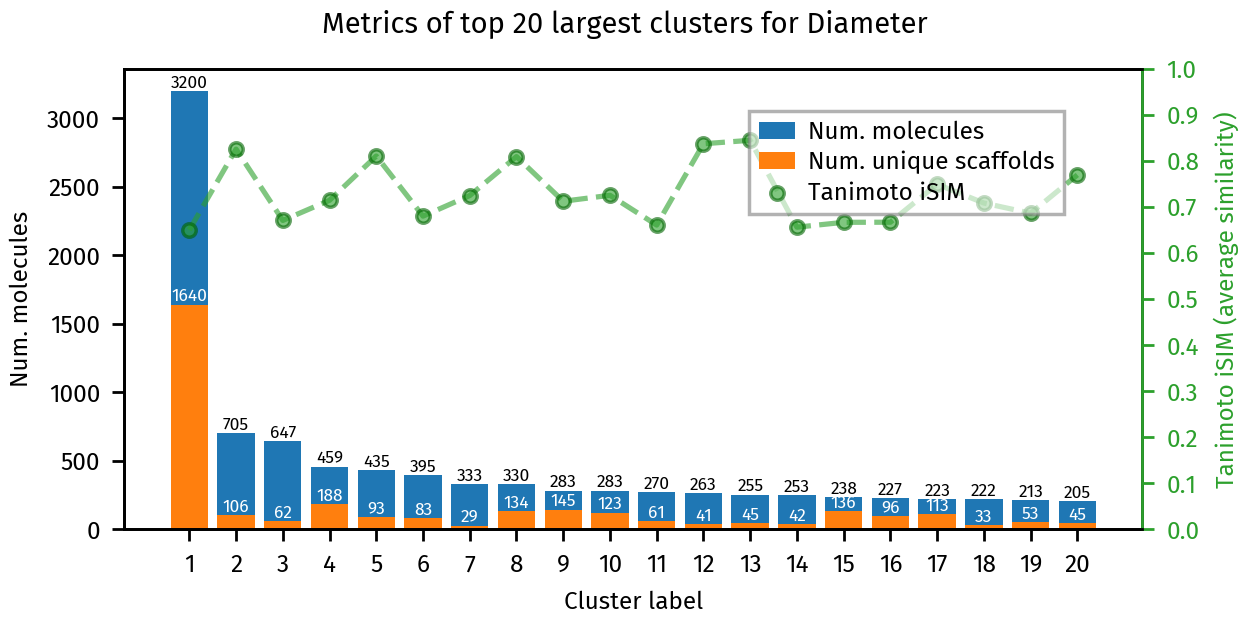
Lets inspect some features of the generated clusters, which we can get directly from the resulting “cluster analaysis”
[54]:
# Total clusters
print("Number of clusters: ", ca.all_clusters_num)
# Clusters with more than 10 molecules
print("Number of clusters with more than 10 molecules: ", ca.all_clusters_num_with_size_above(10))
# Singletons (clusters with a single element)
print("Number of singletons", ca.all_singletons_num)
Number of clusters: 14346
Number of clusters with more than 10 molecules: 964
Number of singletons 8447
Refining the clustering results#
Sometimes you may get a pretty big cluster with a large number of scaffolds, which is undesirable. If this happens, you may want to refine the tree by breaking appart the largest cluster and re-inserting all the bit features. This step is only required if your results display this problem. If they don’t, you are done, no need to refine.
For the refinement, we will use the 'tolerance-diameter' merge criterion, and a tolerance value of 0.0. BitBirch.set_merge(...) can be used to specify the new criterion, and BitBirch.refine_inplace(...) breaks apart the largest cluster and regenerates the tree with our new criterion.
[55]:
# Modify the merge criteria for the tree, from now on bb_tree will use this new criteria
bb_tree.set_merge(merge_criterion="tolerance-diameter", tolerance=0.0)
# Refine the tree using the new merge criteria
bb_tree.refine_inplace(fps)
[55]:
BitBirch(threshold=0.65, branching_factor=50, merge_criterion='tolerance-diameter', tolerance=0.0)
Lets visualize the refined results:
[56]:
clusters_refined = bb_tree.get_cluster_mol_ids()
ca_refined = analysis.cluster_analysis(clusters_refined, fps, smiles)
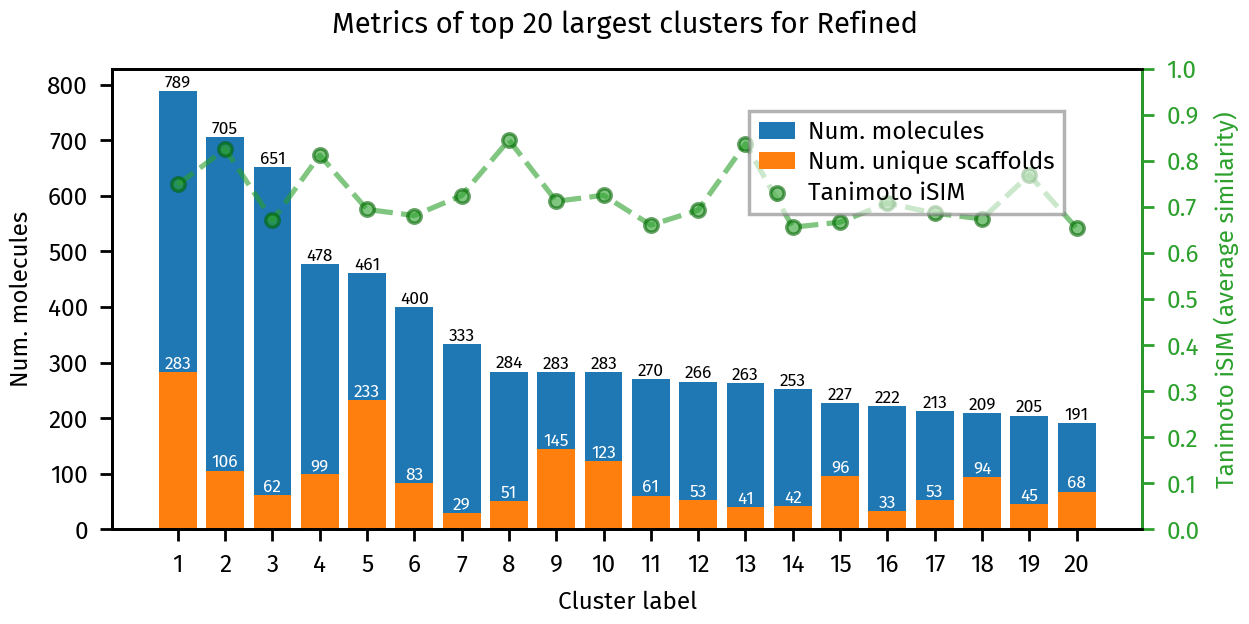
plotting.summary_plot(ca_refined, title="Refined")
# Again, we can save the cluster analysis metrics as a csv file
ca.dump_metrics("./refined-metrics.csv")
plt.show()
[57]:
# Total clusters
print("Number of clusters: ", ca_refined.all_clusters_num)
# Clusters with more than 10 molecules
print("Number of clusters with more than 10 molecules: ", ca_refined.all_clusters_num_with_size_above(10))
# Singletons (clusters with a single element)
print("Number of singletons", ca_refined.all_singletons_num)
Number of clusters: 14735
Number of clusters with more than 10 molecules: 966
Number of singletons 7755
Further analysis and Visualization#
Lets inspect some features of the generated clusters:
We can visually inspect an individual cluster by calling plotting.dump_mol_images. By default this generates multiple images with 30 molecules each.
[58]:
plotting.dump_mol_images(smiles, clusters_refined, cluster_idx=10)
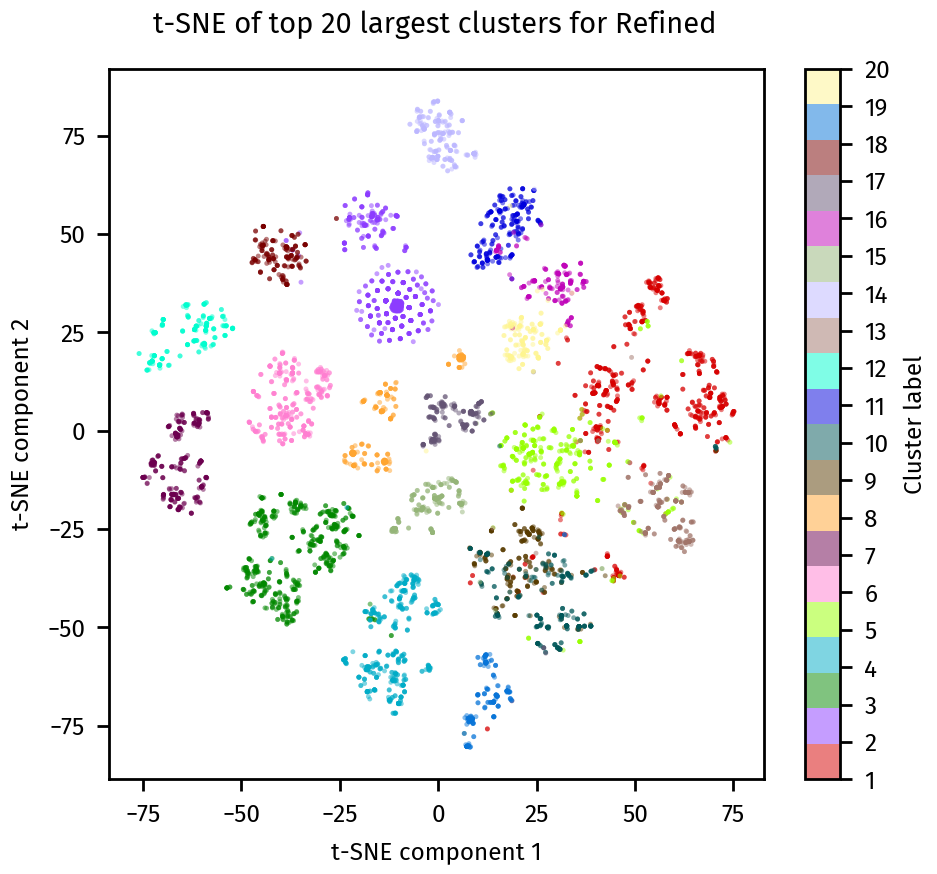
We can also visualize the clusters using a t-SNE plot with plotting.tsne_plot:
[59]:
plotting.tsne_plot(ca_refined, title="Refined")
plt.show()
Once we are happy with the clustering results, we can save the final cluster assignments. to a *.csv file.
[60]:
bb_tree.dump_assignments("smiles-assignments.csv", smiles)